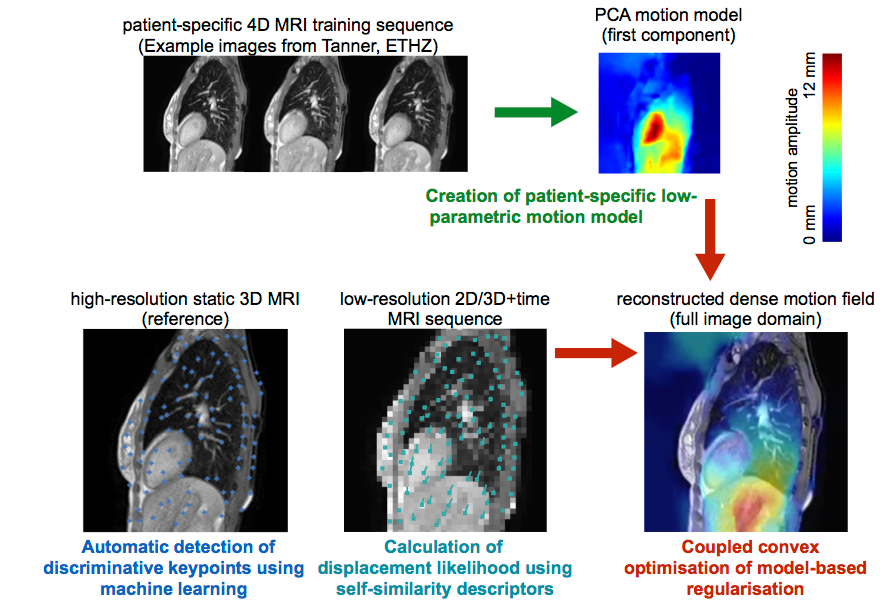

Physiological patient motion is an important problem in accurate dose delivering during radiotherapy. Accurate and realtime motion compensation based on image-guidance could be realised in a combined MR-radiotherapy treatment setup. The objective of this project is to develop algorithms that can estimate intra-fraction motion reliably without implanted fiducial markers and improve on state-of-the-art techniques in terms of accuracy and computational speed.
In contrast to previous work, which predominantly used template matching to achieve realtime speed, we propose to incorporate prior knowledge as well as patient-specific information of plausible deformations during motion estimation. Superior motion estimation, especially for peripheral organs of risk, will be achieved using these models. To account for motion variability, MR images with high temporal resolution acquired during a short setup phase under free breathing could be incorporated for a patient-specific training. Building upon previous work, a motion-model based on principal component analysis or a Bayesian framework can be robustly trained using highly efficient deformable registration. Multiple distributed keypoints at discriminative geometric locations will be extracted automatically using machine learning techniques to avoid the need for invasive implantation of fiducial markers. Robust and accurate realtime motion estimation will be performed within a computationally efficient optimisation framework that incorporates the training model for plausible regularised motion estimation and avoids tracking errors by sampling a large space of potential motion vectors. The algorithms will be validated on retrospective clinical 4D MRI scans using manually annotated landmarks to demonstrate its suitability and advances over state-of-the-art methods.
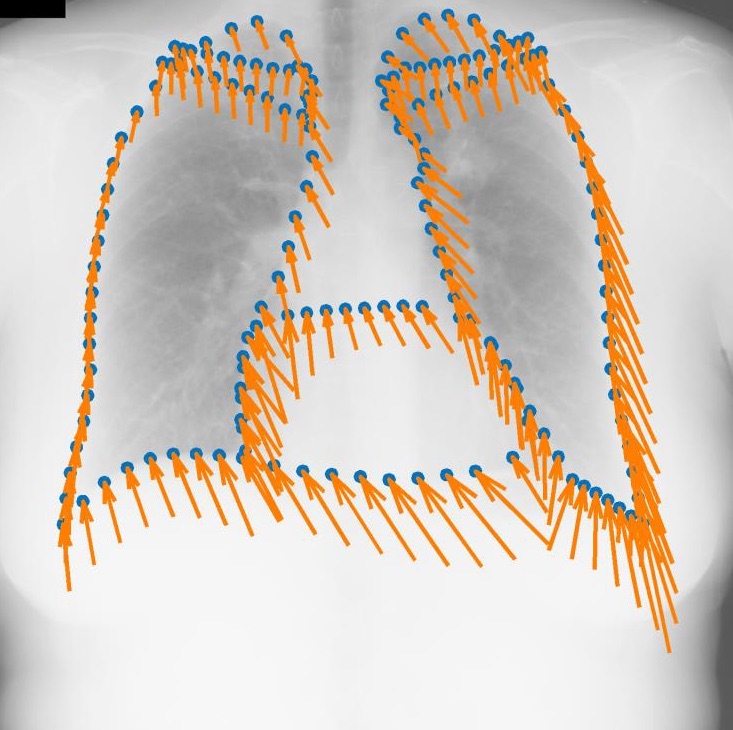
Regression-based Shape Alignment of Multiple Organs: As part of this project, we have adapted the explicit shape regression (ESR) framework of Cao et al. (IJCV 2014) from face alignment to medical image segmentation. Our experiments demonstrate a clear advantage of this approach over commonly used statistical shape models or pixel-wise classification, reducing surface errors from 2.80 to 2.04 mm on the public JSRT dataset. The publication can be found here: Multi-Object Segmentation in Chest X-Ray Using Cascaded Regression Ferns. The dataset is obtainable from Segmentation in Chest Radiographs.
Our MATLAB/Mex implementation is released as open-source code: Shape Regression
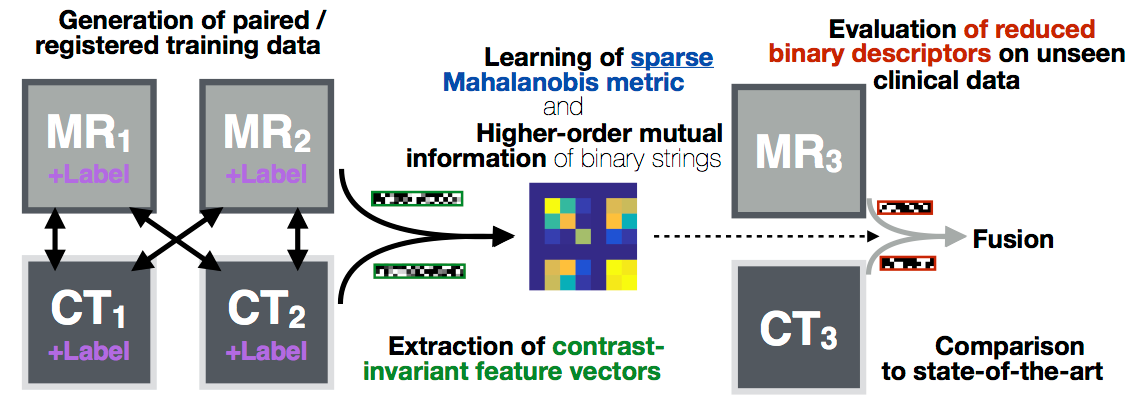
Deformable image registration is a key component for clinical imaging applications involving multi-modal image fusion, estimation of local deformations and image-guided interventions. A particular challenge for establishing correspondences between scans from different modalities: magnetic resonance imaging (MRI), computer tomography (CT) or ultrasound, is the definition of image similarity. Relying directly on intensity differences is not sufficient for most clinical images, which exhibit non-uniform changes in contrast, image noise, intensity distortions, artefacts, and globally non-linear intensity relations (for different modalities).
In this project algorithms with increased robustness for medical image registration will be developed. We will improve on current state-of-the-art similarity measures by combining a larger number of versatile image features using simple local patch or histogram distances. Contrast-invariance and strong discrimination between corresponding and non-matching regions will be reached by capturing contextual information through pair-wise comparisons within an extended spatial neighbourhood of each voxel. Recent advances in machine learning will be used to learn problem-specific binary descriptors in a semi-supervised manner that can improve upon hand-crafted features by including a priori knowledge. Metric learning and higher-order mutual information will be employed for finding mappings between feature vectors across scans in order to reveal new relations among feature dimensions. Employing binary descriptors and sparse feature selection will improve computational efficiency (because it enables the use of the Hamming distance), while maintaining the robustness of the proposed methods.
A deeper understanding of models for image similarity will be reached during the course of this project. The development of new methods for currently challenging (multi-modal) medical image registration problems will open new perspectives of computer-aided applications in clinical practice, including multi-modal diagnosis, modality synthesis, and image-guided interventions or radiotherapy.
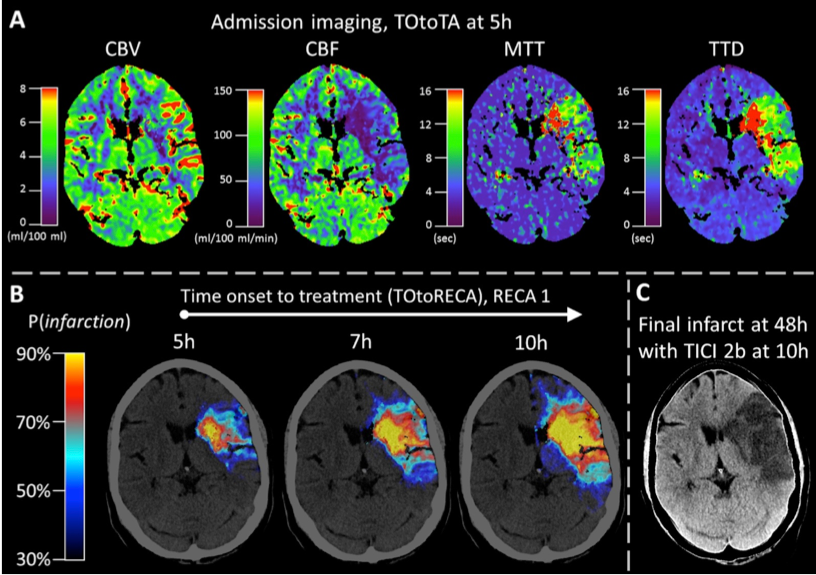
The treatment of acute brain strokes requires very careful decisions in a very narrow time-window. Shortly after patients are admitted to the hospital and scans have been acquired, clinicians need to decide which treatment path offers best chances survival and avoidance of brain damage. These decisions have to be based on a multitude of 3D tomographic image data (CT perfusion maps and potentially thermal imaging) and other clinical indicators (patient age, NIHSS, etc).
Novel image processing techniques, mathematical models and machine learning algorithms, which can deal with the challenges of real clinical data, will be devised, implemented and tested during this research project. A large dataset of retrospective multispectral images of stroke patients is employed for the development and training of new models.
The new algorithms will be used to derive an automatic prediction of a pixel-wise map of tissue that is likely to be recovered if a certain treatment (in particular vessel recanalisation) is performed and how urgent this intervention is. The project is being carried out in close collaboration with the neuroradiology department.
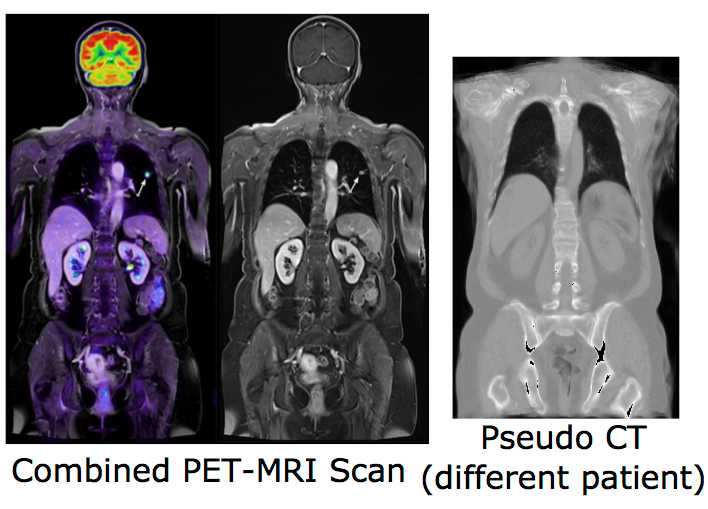
Combining positron emission tomography (PET) and within the same scanner magnetic resonance imaging (MRI) has recently evolved into a research topic of great interest, since this new multimodal imaging technique enables improved tumor localization and delineation from healthy tissue compared to conventional PET-CT. Another important emerging research area is the integration of MRI into radiotherapy treatment delivery systems (e.g. linear accelerators, Linacs) to improve the planning of dose delivery and also tracking and correction of tumor motion. Despite its potential advantages, there are a number of challenging problems when replacing CT completely by MRI in radiotherapeutic treatments and PET imaging, in particular the lack of correlation between the measured MRI intensities and the attenuation-related mass densities. However, since in both cases information about the attenuation behavior of the tissue is required, there is a need for synthetic CT scans (so-called pseudo-CTs) based on the acquired MRI scans.
Our previous work on modality-independent neighbourhood descriptors is considered state-of-the-art for multimodal deformable image registration, which is a vital step for multi-atlas-based CT synthesis. Our first published results for image synthesis include a local formulation of the canonical correlation analysis (CCA) for MRI synthesis based on local histograms and multi-atlas registration based MRI-based pseudo-CT synthesis. In this project, we will advance registration-based techniques and combine them with machine learning algorithms to investigate its potential for image synthesis. For this purpose, various multivariate statistical methods, such as the non-linear Kernel-CCA will be analysed. To overcome the lack of a functional relationship between MRI and CT intensities, the consideration of rich contextual image descriptors will be studied. Learning algorithms for detecting and correcting errors in the generation of pseudo-CT scan could also be explored. Following image synthesis, attenuation maps can be directly created based on the synthesized pseudo-CT images and its influence on PET image reconstruction may be evaluated. Due to the limitation of previous work on the generation of attenuation maps mainly for the head, a particular focus of this project is the development of new methods for MRI-based attenuation correction for the whole body.
Supplementary data for COPD registration paper:
The manual segmentations of oblique fissures of 10 inhale/exhale of the DIR-Lab COPD dataset are available for download to evaluate fissure distances: FissuresUpload.zip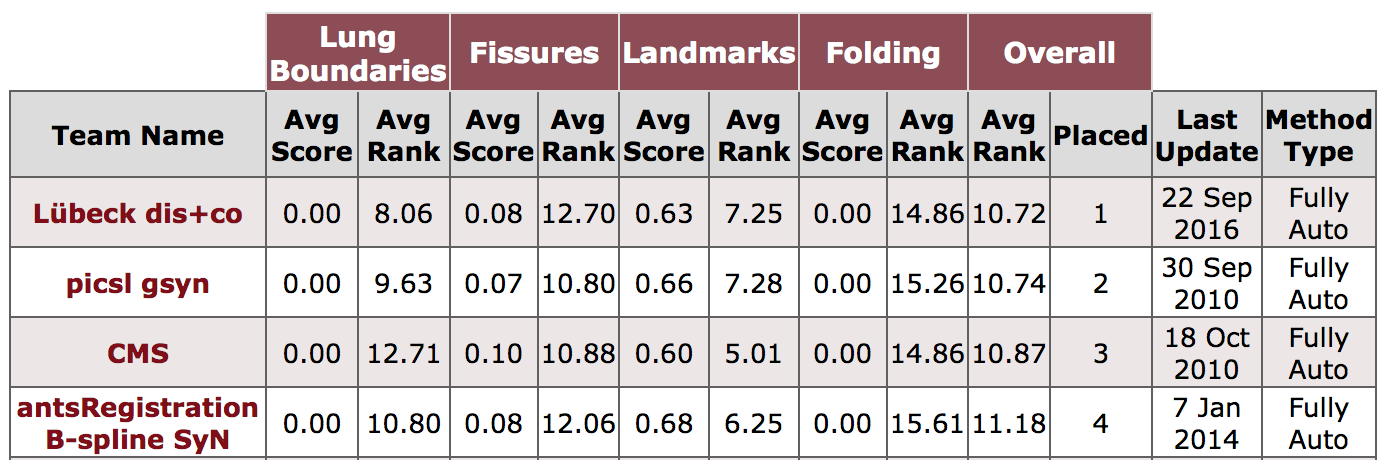
Federated Training and Inference for Learning Based and GPU-Accelerated 3D Registration Methods:
3D registration is an important area in medical imaging that (e.g. in contrast to medical segmentation) has yet to fully benefit from deep learning and GPU computing. We want to work towards sustainable and fair research in medical image registration by exploring new solutions for federated training of DL-based 3D registration and enabling others to leverage large-scale hidden medical datasets with privacy preservation. We also plan to provide GPU-accelerated implementations of cutting-edge non-DL registration frameworks (see references below) to be used with external data e.g. by clinical researchers or low-income institutions. This will enable direct impact for a multitude of medical tasks. For the ongoing MICCAI 2021 Learn2Reg challenge please visit: https://learn2reg.grand-challenge.org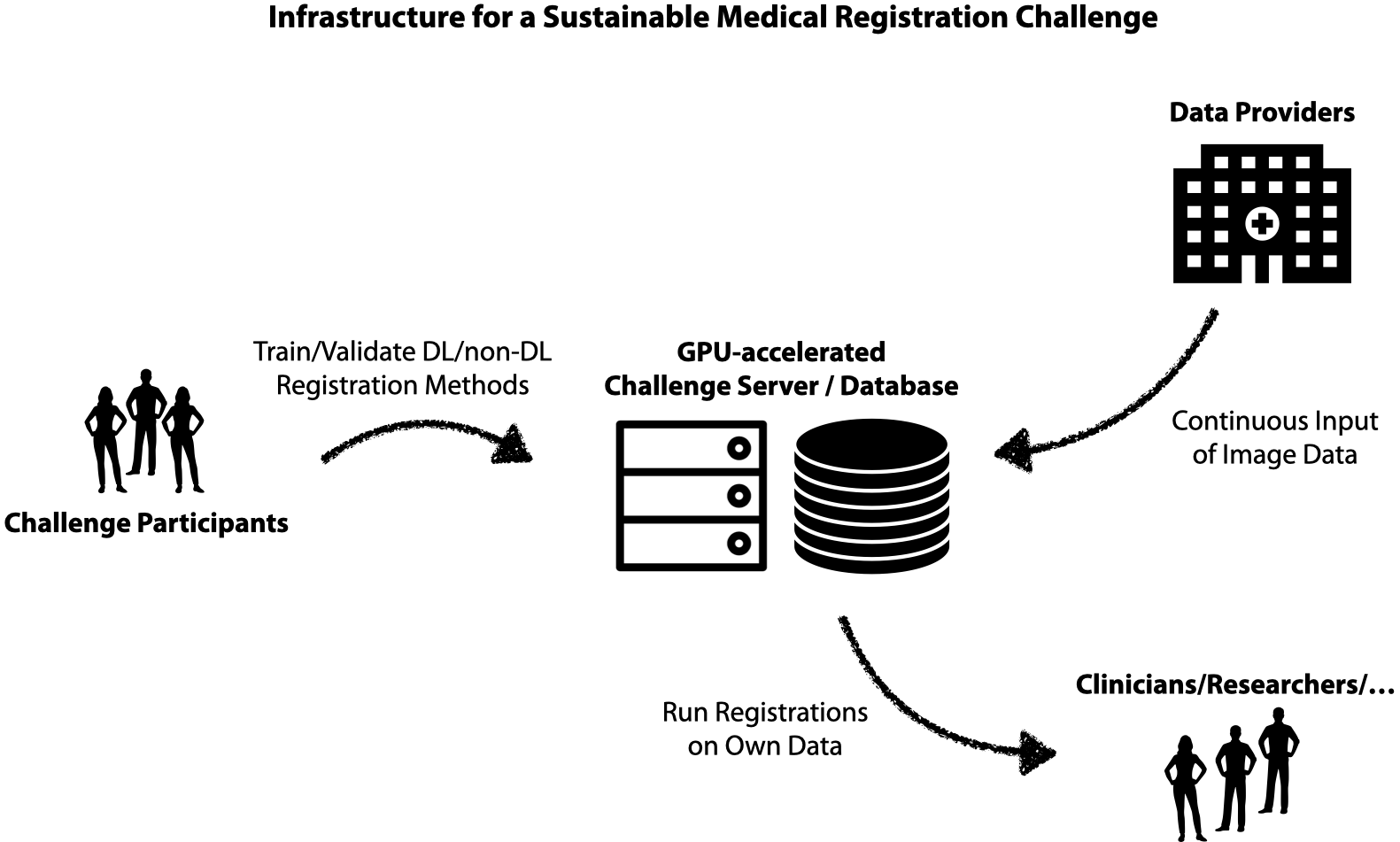
Vantage Point Forest (MICCAI 2016):
Code for the training and application of the vantage point forests can be found here: Matlab/Mex ZipThe following video demonstrates the accurate segmentation obtained using our VP forest for abdominal organs in 3D CT. The data is obtained from VISCERAL.eu. Applying a trained model to an unseen scan takes less than 2 seconds.